
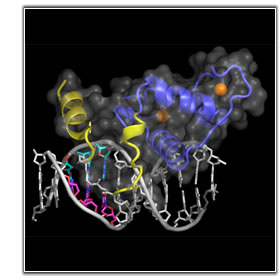
 The central methodology employed in our laboratory is macromolecular crystallography. Additional structural approaches we have utilized include NMR and Atomic Force Microscopy.
The central methodology employed in our laboratory is macromolecular crystallography. Additional structural approaches we have utilized include NMR and Atomic Force Microscopy.
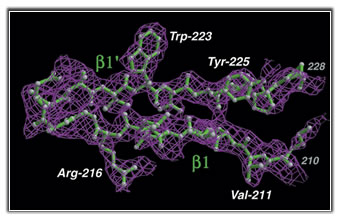
We use the structural information we collect to enhance biological discovery. The data is also applicable to the chemical biology, protein design and nanotechnology approaches pursued by members of our lab.
A representative set of publications is provided below:
Teotico, D.G., Bischof, J.J., Peng, L., Kliewer, S.A., and Redinbo, M.R. (2008).
Structural basis of PXR activation by the hops constituent colupulone.
Molecular Pharmacology, 74, 1512-1520.
Schaaf, G., Ortlund, E.A., Tyeryar, K., Mousley, C., Ile, K., Garret, T., Ren, J., Woolls, M., Raetz, C.R.H., Redinbo, M.R., and Bankaitis, V. A. (2008).
Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the Sec14-superfamily.
Molecular Cell, 29, 191-206.
Thapar, R., Marzluff, W.F., and Redinbo, M.R. (2004).
Electrostatic contribution of serine phosphorylation to the Drosophila SLBP-histone mRNA complex.
Biochemistry, 43, 9401-9412.
Xue, Y., Ratcliff, G.C., Wang, H., Davis-Searles, P.R., Gray, M.D., Erie, D.A., and Redinbo, M.R. (2002).
A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3’-5’ exonuclease and 5’-protruding strand endonuclease activities.
Biochemistry, 41, 2901-2912.